Research
Our Main Research Areas
Our research interests are centered in two main areas:
- mechanistic study of transcription factors (TFs) for plant metabolic pathway regulation, and
- improvement of bioenergy production through engineering tobacco plants to produce high-density biofuels and engineering enzymes for novel functions in biomass conversion.
A new research focus in our lab is the modification of leaf characteristics and surface chemistry. The research in our lab addresses some of the key issues in molecular biology and protein biochemistry and contributes to the development of new-crop opportunities and bioenergy, two important areas in the agricultural economies of Kentucky and the nation.
Structure/function Relationship and Regulation of Plant Transcription Factors
TFs are regulatory proteins that modulate gene transcription. Our approach to understanding the regulatory mechanism relies on dissection of the structure/function relationships of TFs involved in regulating plant secondary pathways. We are particularly interested in two families of TFs, the basic helix-loop-helix (bHLH) and MYB TFs, that are involved in several well-known plant metabolic pathways including the flavonoid biosynthetic pathway. These TFs represent two large TF superfamilies in diverse eukaryotic organisms ranging from yeast to plants and animals. The combinatorial regulation of gene expression by the bHLH/MYB complex has been established as an excellent model for understanding plant metabolic pathway control. However, the structure/function basis and the combinatorial regulatory mechanism of the bHLH/MYB TFs are not well understood.
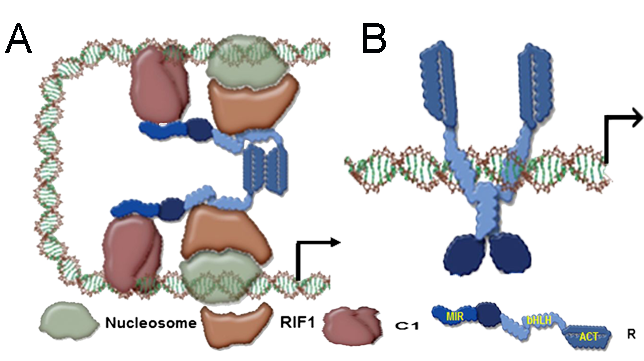
Proposed model for promoter switching by R. (A) ACT domain ON model shows C1 making the DNA contacts and recruiting R through its N-terminal MYB-interacting region (MIR). The ACT C-terminal region of R forms a dimer; therefore the bHLH motif remains as a monomer and can bind to RIF1, which recognizes chromatin components. This model explains the expression of A1. (B) ACT domain OFF model shows R binding to DNA by forming homodimers through the bHLH because the ACT is not homodimerizing. C1 continues to interact with the MIR region of R, providing transcriptional activatory function.
Many questions remain in our understanding of the combinatorial transcriptional regulation mediated by bHLH/MYB TFs. For example, how are pathway genes that lack obvious conserved cis-regulatory elements regulated by the same set of TFs? Also, as cellular levels of TF proteins increase, how do TFs maintain a delicate balance in vivo without causing hyperactivation? My lab addresses some of these difficult questions by examining the structure/function relationship, gene expression patterns, DNA-binding ability, protein-protein interactions and posttranslational modification of these TFs. We uncovered a mechanism by which a dimerization domain in a bHLH TF behaves as a switch that permits different configurations of TF regulatory complexes to be tethered to different promoters, helping to explain the elusive question of how genes lacking conserved cis-elements in their promoters can be coordinately regulated. We also demonstrated that bHLH TFs regulating flavonoid biosynthesis are posttranslationally modulated by the 26S proteasome in Arabidopsis.
Transcriptional Regulation of Terpenoid Indole Alkaloid Biosynthesis
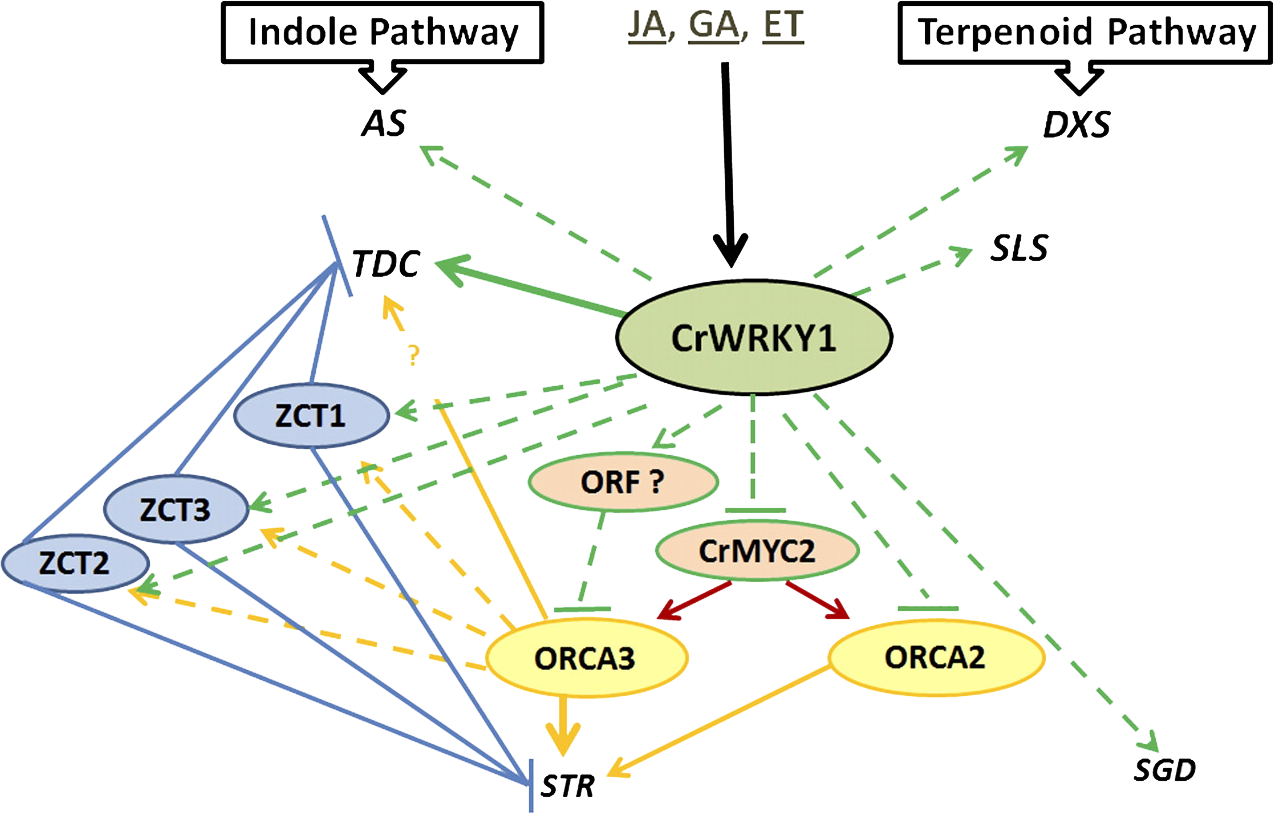 We are also interested in transcriptional control of a metabolic pathway with pharmaceutical importance. The medicinal plant Madagascar periwinkle (Catharanthus roseus) produces a large array of terpenoid indole alkaloids (TIAs), an important source of natural or semisynthetic anticancer drugs. However, the transcriptional regulation of the TIA pathway is poorly understood. We utilized various genomic tools and discovered that WRKY TFs are involved in regulation of TIA biosynthesis. WRKY TFs form a large TF family and play dynamic roles in, among other biological processes, biotic and abiotic stress responses. We identified and functionally characterized the first C. roseus WRKY TF for its role in TIA biosynthesis. We perform comprehensive bioinformatic and gene ontology analysis of all WRKY TFs in C. roseus, and experimentally verify their predicted functions. We are also investigating how these WRKY TFs are regulated in various signal transduction pathways and their relationship with other TFs.
We are also interested in transcriptional control of a metabolic pathway with pharmaceutical importance. The medicinal plant Madagascar periwinkle (Catharanthus roseus) produces a large array of terpenoid indole alkaloids (TIAs), an important source of natural or semisynthetic anticancer drugs. However, the transcriptional regulation of the TIA pathway is poorly understood. We utilized various genomic tools and discovered that WRKY TFs are involved in regulation of TIA biosynthesis. WRKY TFs form a large TF family and play dynamic roles in, among other biological processes, biotic and abiotic stress responses. We identified and functionally characterized the first C. roseus WRKY TF for its role in TIA biosynthesis. We perform comprehensive bioinformatic and gene ontology analysis of all WRKY TFs in C. roseus, and experimentally verify their predicted functions. We are also investigating how these WRKY TFs are regulated in various signal transduction pathways and their relationship with other TFs.
Improvement of Bioenergy Production
Another research focus of our lab is bioenergy production. Two particular focuses have been on
- engineering tobacco as a production platform for high-density biofuels
- protein engineering of cellulosic hydrolases.
 Currently we are a member of FOLIUM (Tobacco as a platform for foliar biosynthesis of advanced hydrocarbon fuels), a project supported by DOE (ARPA-E). This multi-institutional collaborative project involving Lawrence Berkeley National Lab, UC Berkeley and UK aims at producing high-density liquid fuels in green biomass by introducing genes from microorganisms and other plants, resulting in accumulation of hydrocarbon fuels in tobacco leaves and stems. The FOLIUM project is newsworthy and was featured by the New York Times and the Los Angeles Times . To learn more about FOLIUM please visit here.
Currently we are a member of FOLIUM (Tobacco as a platform for foliar biosynthesis of advanced hydrocarbon fuels), a project supported by DOE (ARPA-E). This multi-institutional collaborative project involving Lawrence Berkeley National Lab, UC Berkeley and UK aims at producing high-density liquid fuels in green biomass by introducing genes from microorganisms and other plants, resulting in accumulation of hydrocarbon fuels in tobacco leaves and stems. The FOLIUM project is newsworthy and was featured by the New York Times and the Los Angeles Times . To learn more about FOLIUM please visit here.
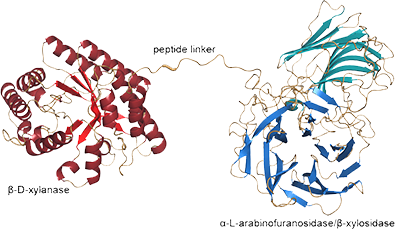 The other research area is the creation of multi-functional enzymes for lignocellulosic biomass conversion. Our approaches include the application of directed enzyme evolution and the creation of artificial multidomain enzymes. Several of the engineered enzymes display synergistic effects that lead to higher efficiencies in xylan-degradation compared to the parental enzyme mixtures. Our results demonstrate the feasibility and advantages of the multifunctional enzyme strategy for reducing the total number of proteins required in biomass conversion. This research has been recognized as novel and groundbreaking by other leading researchers working on related designer enzyme approaches to biomass deconstruction [Appl. Environ. Microbiol. (2010)76: 3787]. In addition, we continue to work in collaboration with two USDA labs to apply directed evolution technologies to improve cellulosic hydrolases, particularly in properties related to product inhibition and thermostability.
The other research area is the creation of multi-functional enzymes for lignocellulosic biomass conversion. Our approaches include the application of directed enzyme evolution and the creation of artificial multidomain enzymes. Several of the engineered enzymes display synergistic effects that lead to higher efficiencies in xylan-degradation compared to the parental enzyme mixtures. Our results demonstrate the feasibility and advantages of the multifunctional enzyme strategy for reducing the total number of proteins required in biomass conversion. This research has been recognized as novel and groundbreaking by other leading researchers working on related designer enzyme approaches to biomass deconstruction [Appl. Environ. Microbiol. (2010)76: 3787]. In addition, we continue to work in collaboration with two USDA labs to apply directed evolution technologies to improve cellulosic hydrolases, particularly in properties related to product inhibition and thermostability.
Collaborators

Professor Erich Grotewold |

Dr. Xuguo Zhou University of Kentucky |

Dr. Kurt Wagschal |

Transposagen Biopharmaceuticals, Inc. |
Funding Agencies
 |
 |
 |
 |
 |
 |
